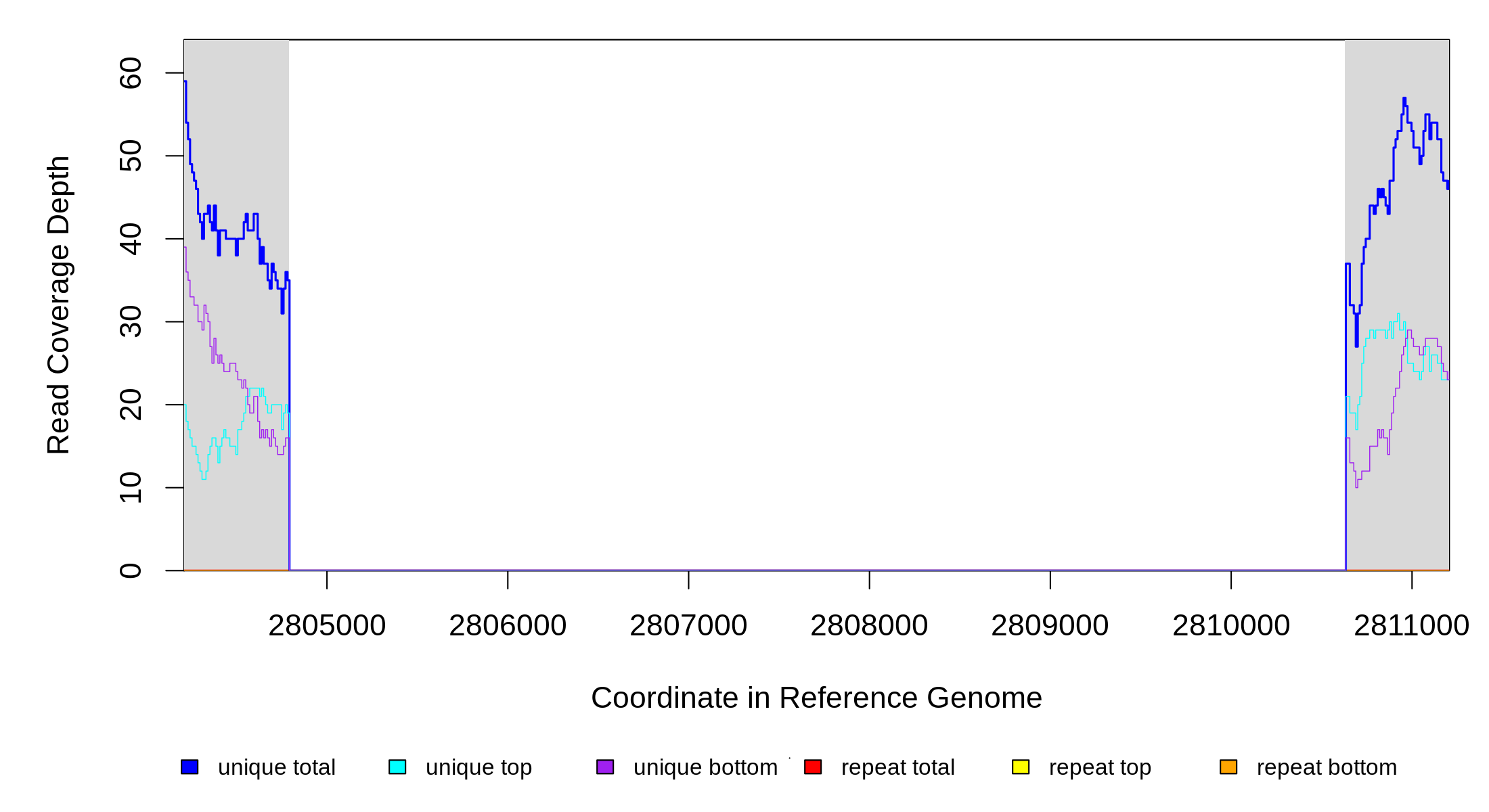
| Predicted mutation | ||||||
|---|---|---|---|---|---|---|
| evidence | seq id | position | mutation | annotation | gene | description |
| MC JC | NC_000913 | 2,804,790 | Δ5,840 bp | proV–[ygaH] | proV, proW, proX, ygaZ, [ygaH] | |
| Missing coverage evidence... | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| seq id | start | end | size | ←reads | reads→ | gene | description | |||
| * | * | ÷ | NC_000913 | 2804790 | 2810629 | 5840 | 35 [0] | [0] 35 | proV–[ygaH] | proV,proW,proX,ygaZ,[ygaH] |
| New junction evidence | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| seq id | position | reads (cov) | reads (cov) | score | skew | freq | annotation | gene | product | ||
| * | ? | NC_000913 | = 2804789 | 0 (0.000) | 35 (0.570) | 34/492 | 0.4 | 100% | intergenic (+328/‑26) | nrdF/proV | ribonucleoside‑diphosphate reductase 2 subunit beta/glycine betaine ABC transporter ATP binding subunit ProV |
| ? | NC_000913 | 2810630 = | 0 (0.000) | coding (287/336 nt) | ygaH | L‑valine exporter, YgaH component | |||||
