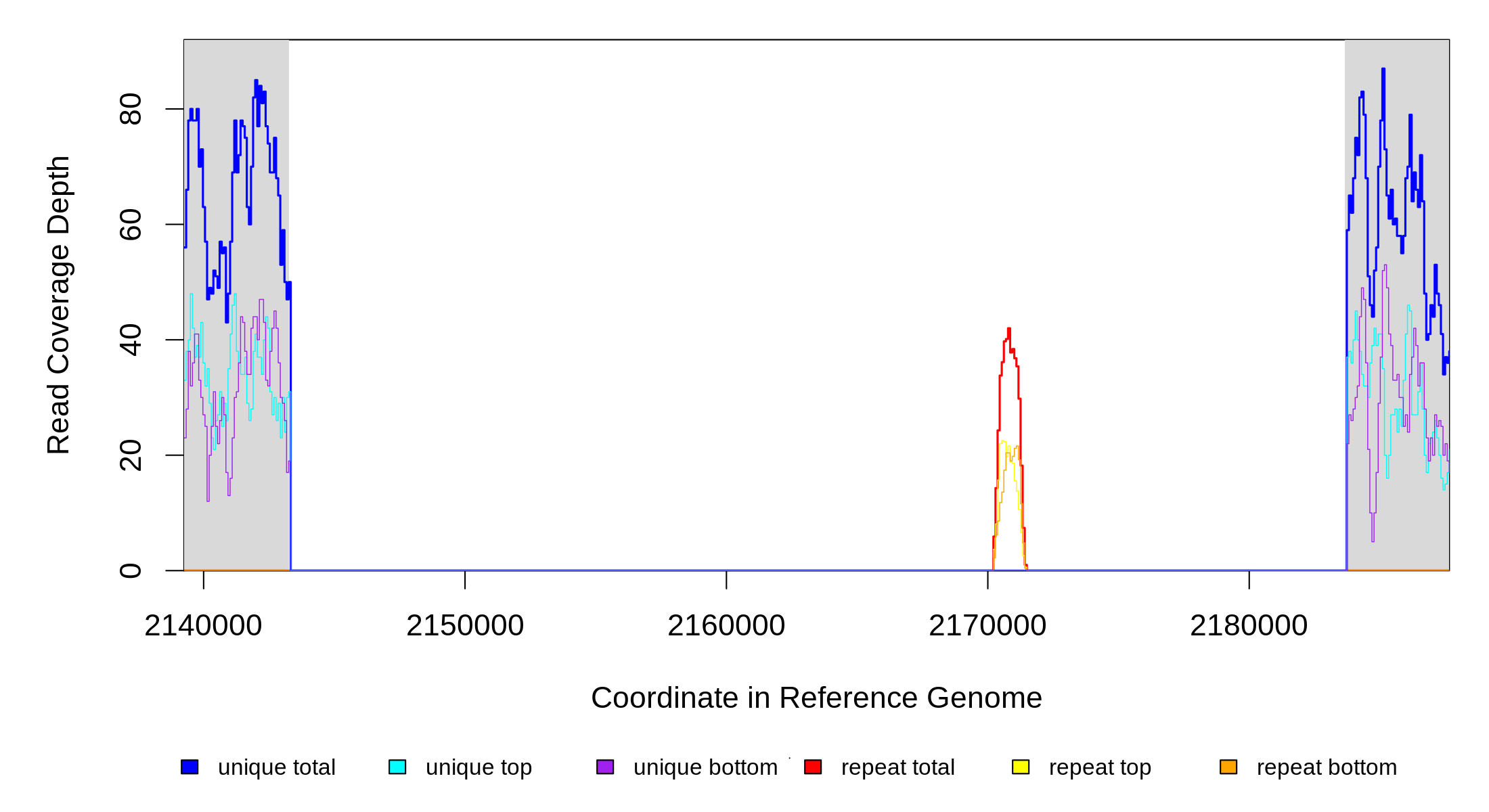
| Predicted mutation | ||||||
|---|---|---|---|---|---|---|
| evidence | seq id | position | mutation | annotation | gene | description |
| MC JC | NC_000913 | 2,143,266 | Δ40,395 bp | [dgcE]–yegX | 38 genes [dgcE], alkA, yegD, yegI, yegJ, pphC, yegL, sibA, ibsA, sibB, ibsB, mdtA, mdtB, mdtC, mdtD, baeS, baeR, yegP, trhP, cyaR, ogrK, yegR, yegS, gatR, insE5, insF5, gatR, gatD, gatB, gatA, gatZ, gatY, fbaB, yegT, yegU, yegV, yegW, yegX |
|
| Missing coverage evidence... | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| seq id | start | end | size | ←reads | reads→ | gene | description | |||
| * | * | ÷ | NC_000913 | 2143266 | 2183660 | 40395 | 50 [0] | [0] 51 | [dgcE]–yegX | [dgcE],alkA,yegD,yegI,yegJ,pphC,yegL,sibA,ibsA,sibB,ibsB,mdtA,mdtB,mdtC,mdtD,baeS,baeR,yegP,trhP,cyaR,ogrK,yegR,yegS,gatR,insE5,insF5,gatR,gatD,gatB,gatA,gatZ,gatY,fbaB,yegT,yegU,yegV,yegW,yegX |
| New junction evidence | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| seq id | position | reads (cov) | reads (cov) | score | skew | freq | annotation | gene | product | ||
| * | ? | NC_000913 | = 2143265 | 0 (0.000) | 50 (0.820) | 40/492 | 0.2 | 100% | intergenic (‑317/‑1) | udk/dgcE | uridine/cytidine kinase/putative diguanylate cyclase DgcE |
| ? | NC_000913 | 2183661 = | 0 (0.000) | intergenic (‑10/+55) | yegX/thiD | putative glycosyl hydrolase YegX/bifunctional hydroxymethylpyrimidine kinase/phosphomethylpyrimidine kinase | |||||
