|
■ Bioresources information is available at the following URLs
|
 |
|
Research and Bioresources <No. 29> Phenotype ontology for further use of bioresources
Hiroshi Masuya (Unit Leader, Technology and Development Unit for Knowledge Base of Mouse Phenotype, RIKEN BioResource Center)
"Phenotype" is the set of properties or characteristics of an organism that result from its "genotype." For example, "phenotype" is expressed by a combination of "traits," such as flower color, seed shape, eye color, and body weight of an animal and specific “characteristics (= values),” such as pink (flower), cockle (seed), red (eye), and 90 kg (animal). When selecting a bioresource for use in research, the "phenotype" is one of the important sources of information, similar to the "genotype." However, since the "phenotype" is not systematically described in many cases, it has been difficult to use the information on "phenotype" effectively in a database. Recently, because of the advances in the technology of ontology, which can handle a complicated concept in a database, "phenotype" can now be examined as a conceptual structure. Thus, "phenotype" can be linked with "disease information." In this article, I would like to introduce such trends. |
In 1995, gene ontology (GO) was proposed for the first time in the field of bioinformatics. This methodology was introduced in the context of phenotype as phenotype ontology. In the past, when compiling a phenotype database, the text explaining a phenotype was digitized, and a full-text search was performed. In the database, a major problem encountered was "notation variability." For example, when searching for a mouse exhibiting the falling-out of hair phenotype, various expressions, such as hairless, bald, nude, alopecia, and hair loss were used in each text explaining a phenotype. Therefore, an exhaustive search could not be performed. Phenotype ontology was then created in a manner by which concepts were separated from labels, synonyms were summarized, semantic similarity was indicated by linking identifications, and terms and their meanings were summarized in the form of a database.
The Mammalian Phenotype Ontology (MP) was the first phenotype ontology created in the Jackson Laboratory's Mouse Genome Informatics (MGI) in order to exhibit the characteristics of mouse strains as bioresources (Ref. 1). The MGI abolished the full-text search for phenotype information, and around 2005, the mouse strains were summarized according to the MP. As a result, the existing mouse strains could be classified using phenotypes, and mouse strains with similar phenotypes could be searched exhaustively. Thus, the MGI has greatly contributed to research in the post-genome era. |
The terms and concepts used to indicate phenotypes differ slightly among the research communities studying specific species. Therefore, different ontologies have been created in different research fields, such as Human Phenotype Ontology (HPO) (Ref. 2), Zebrafish Phenotype Ontology (ZPO) (Ref. 3), and Worm Phenotype Ontology (WP) (Ref. 4). Moreover, by manual curation and mechanical inference, phenotype terms could be linked across species (Fig. 1) and the data on cross-species phenotype terms are open to the public. In particular, since the HPO is linked with the Online Mendelian Inheritance in Man (OMIM) and Disease Ontology (DO), the HPO can demonstrate the relationship between a disease and a phenotype (Ref. 5).
|
|
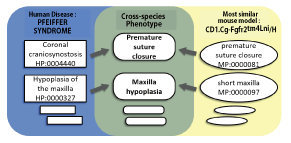
Fig. 1. Linkage among cross-species phenotype terms (Ref. 5 was amended)
Regarding the disease shown on the left, the overall characteristics of the morbid state can be exhibited by multiple HPO terms. Similarly, the overall characteristics of the mouse strain shown on the right can be exhibited by multiple MP terms. By linking with the cross-species phenotype terms shown in the center, phenotypes of different species can be matched and the relationship between the disease and the animal model can be demonstrated. (A sketch obtained at
http://genome.cshlp.org/content/24/2/340/F2.expansion.html was amended)
|
|
Disease information and phenotype ontology |
The bioresources of vertebrates play an important role as animal disease models. Here, the trend of ontology in the medical field has been examined. Since sharing of information is an important issue in the medical field, to create a list of standard disease names and to widely share the created list have been attempted. Recently, in addition to the list of standard disease names, the HPO has attracted much attention. In the medical field, phenotypes can correspond to "morbid state" and "test results," which are more precise than disease names. A disease name is determined by a doctor's diagnosis. However, when the HPO is used, information on the morbid state or the test results of a disease, which are the basis of diagnosis, can be shared by many doctors. For example, some of the genetic and rare diseases are called “rare and undiagnosed diseases.” Regarding rare and undiagnosed diseases, the disease name cannot be identified even though the symptom clearly appears, as the number of cases is small. As a result, development of the therapeutic method is delayed.
If the gene causing a rare and undiagnosed disease is identified, development of the therapeutic method can be greatly advanced. Since information sharing at the level of disease name cannot utilize the information on patients suffering from rare and undiagnosed diseases, the gene causing a rare and undiagnosed disease is difficult to identify. Information sharing using the HPO provides an opportunity to identify the causative gene. At present, the Japan Agency for Medical Research and Development has been constructing a nationwide information network of medical institutions related to rare and undiagnosed diseases using the HPO through the Initiative on Rare and Undiagnosed Diseases (6). |
Role of phenotype ontology in the effective use of bioresources |
Similar to the above-mentioned example of rare and undiagnosed diseases, experimental animals, that are homologous with diseases at the phenotype level, contribute greatly to research and development as disease models. As mentioned above, bioresource-related institutions have been developing the foundation for information technology that handles phenotype data. As a result, data with the following linkage can be used: disease → morbid state (human phenotype) → model organism phenotype → genotype → strain. Recently, ontology has been constructed based on data sharing through the Internet, the internationally standardized technology to be used commonly in databases and the Resource Description Framework (RDF). Similar to the above-mentioned linkage of data, the movement to link the bioresources worldwide with the related information has been accelerated. |
In Japan, the National BioScience Database Center (NBDC) has been promoting data sharing based on the RDF. Our research group has been operating the "J-phenome" (7), a Database Integration Coordination Program in the NBDC, in which the phenotype data on bioresources in Japan are integrated using the RDF. In cooperation with institutions providing the bioresources of mouse, rat, medaka, zebrafish, etc., the J-phenome has used ontology to annotate phenotype information, used the RDF to integrate data, and released the original data of each bioresource-providing institution in the form of “metadata,” which further explains the original data using an internationally standardized method. The J-phenome has also begun sharing information with the Monarch Initiative (8), a worldwide phenotype data integration project, and the International Mouse Phenotyping Consortium (9). Through these activities, the J-phenome would contribute to expanding the effective use of bioresources in Japan (Fig. 2). The J-phenome will soon release a system, by which each bioresource can be searched using disease names, phenotypes, and organs, tissues, or genes exhibiting the phenotype through the above-mentioned linkage of data. Please use the system for your research. |
|
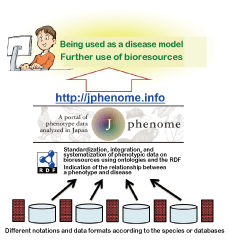
Fig. 2. Conceptual diagram of the J-phenome
In cooperation with the domestic and foreign databases related to phenotypes and bioresources, the J-phenome collects phenotype data. The differences in notation and data formats are solved by the semantic web technology using ontologies and the RDF. The original data is released in the form of "metadata," which further explains the original data. Through these activities, the J-phenome aims to increase the efficiency of using bioresources and phenotype data.
|
|
Ref. 1. |
Smith CL et al. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 2005;6(1):R7 |
Ref. 2. |
Köhler S et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014 Jan;42:D966-74.
|
Ref. 3. |
Robinson PN, Webber C. Phenotype ontologies and cross-species analysis for translational research. PLoS Genet. 2014 Apr 3;10(4):e1004268. |
Ref. 4. |
Schindelman G et al. Worm Phenotype Ontology: integrating phenotype data within and beyond the C. elegans community. BMC Bioinformatics. 2011 Jan 24;12:32. |
Ref. 5. |
Robinson PN et al. Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome Res. 2014 Feb;24(2): 340-8. |
(6) |
http://www.amed.go.jp/program/IRUD/
|
(7) |
http://jphenome.info |
(8) |
https://monarchinitiative.org |
(9) |
http://www.mousephenotype.org |
|
|
|
Linux tools can now be run on Windows 10 ! Part 2 |
Continuing from the previous edition, this is the second part of the article introducing how to install and use Linux tools on Windows 10.

Software executed from the bash command line can either be installed using the apt package management tool or compiled from source code. As an example, the procedure for installing TopHat, a tool used in RNA-Seq mapping, is described below.
You must first install dependent libraries before compiling TopHat. These libraries can be installed using the apt-get command (Fig. 1).
[ sudo apt-get install –y g++ libboost-all-dev make ]
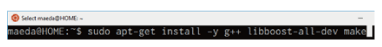
Fig. 1. Installing software using apt-get
Once the required libraries have been installed,
TopHat (https://ccb.jhu.edu/software/tophat/index.shtml) can be compiled from its source code. Run the following commands:
[ curl -XGET https://ccb.jhu.edu/software/tophat/downloads/tophat-2.1.1.tar.gz > tophat-2.1.1.tar.gz]
[ tar xvzf tophat-2.1.1.tar.gz ]
[ cd tophat-2.1.1 ]
[ sudo ./configure ]
[ sudo make ]
[ sudo make install ]
If you are asked to enter a password, use the one that you configured during the installation of Bash on Ubuntu on Windows. After the compilation and installation are finished, type "tophat" and press the [Enter] key. If a message like the following is displayed, then the installation was successful (Fig. 2).
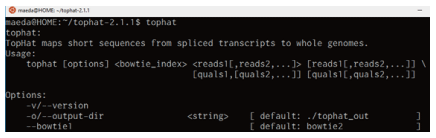
Fig. 2. Running TopHat (showing the command usage)

Accessing Windows files from Bash on Ubuntu on Windows requires some care. For example, the path for accessing the D drive is [/mnt/d], and backslash [\] must be replaced with a forward slash [/]. For example, the file
[ D:\Downloads\tophat-2.1.1.tar.gz ]
can be accessed using this path:
[ /mnt/d/Downloads/tophat-2.1.1.tar.gz ]
from Bash on Ubuntu on Windows.
The author has successfully installed various tools using Bash on Ubuntu on Windows in Windows 10 using apt-get, as well as by compiling from the source code. Until now, many people have preferred to use Mac to use command line tools. Now that this has also become possible in Windows, there is a wider range of hardware to choose from when purchasing a new computer.
|
|
|
(Shunsuke Maeda) |
|