|
 |
■Bioresources information is available at the following URLs)
|
 |
|
Introduction to Resource Center <No. 56> Specialty of Human Cord Blood Stem Cell Resources for Research in the National BioResource Project
Tokiko Nagamura, Associate Professor (Dept. of Cell Processing and Transfusion,
Research Hospital, The Institute of Medical Science, The University of Tokyo)
Human cord blood stem cells are derived from the blood of babies who were normal and healthy at birth. Human cord blood stem cells are embryonic cells that contain abundant undifferentiated hematopoietic stem cells and tissue stem cells, such as immature immune cells and mesenchymal stem cells. Therefore, human cord blood stem cells have been widely used for medical and biological sciences, including research in the areas of regenerative medicine, drug discovery, cancer, immunology, infectious diseases, genetics, and environmental medicine. In particular, human cord blood stem cells are considered to be bioresources that are essential for research on the life sciences. Clinically, cord blood has been studied as a resource for hematopoietic stem cell transplantation for various types of serious blood dyscrasia, congenital immunodeficiency, and metabolic disease, together with the bone marrow and peripheral blood stem cells. In Japan, more than 1,000 patients a year are treated by cord blood stem cell transplantation (this is the most of any country in the world!).
Public cord blood banks for clinical use have been established to support cord blood stem cell transplantation. In this system, cord blood collected in affiliated obstetrics and gynecology departments is transported to public cord blood banks for clinical use. The transported cord blood is prepared and stored in the banks, and the stored cord blood is provided as needed for patients who need cord blood stem cell transplantation. However, public cord blood banks for clinical use only prepare and store cord blood that contains a large number of cells, which is commonly utilized clinically. Therefore, 70%−80% of the collected cord blood is not adopted for clinical use. Some of the unadoptable cord blood is used for research or for examining the cord blood banking system, and the rest is discarded.
|
The Research Cord Blood Stem Cells Bank of the National BioResource Project (NBRP) supported by the Ministry of Education, Culture, Sports, Science and Technology was established to collect, prepare, and cryopreserve cord blood that does not satisfy the criteria for clinical use in public cord blood banks, and to deliver such cord blood to foreign and domestic researchers, who perform research for the development of medical science, through the RIKEN BioResource Center (RIKEN BRC). Fig. 1 shows the flow of cord blood in the Research Cord Blood Stem Cells Bank project system. |
|
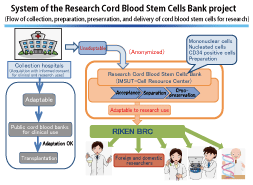
Figure 1. Flow of cord blood delivery |
|
Types and Characteristics of Cells for Delivery |
| There are large differences between the type of samples handled by the Research Cord Blood Stem Cells Bank and those handled by other organizations. The samples handled by the Research Cord Blood Stem Cells Bank are human primary cells that are discarded after use, and that have been prepared and cryopreserved at the Research Cord Blood Stem Cells Bank. As shown in Table 1, the Research Cord Blood Stem Cells Bank delivers several types of cell samples. Researchers can select the cell type according to their research purpose.
|
|

Table 1. Types of human cord blood samples for research
|
|
| Nucleated cell samples contain all of the types of white blood cells in cord blood. In other words, the samples contain hematopoietic stem cells (CD34 positive cells), mononuclear leukocytes (consist mainly of lymphocytes and monocytes), and neutrophils. A reagent called hydroxyethyl starch (HES) is added to cord blood, which polymerizes with red blood cells in order to remove as many red blood cells as possible. The remaining types of cells are collected and concentrated. A cryoprotective agent is added to aid in freezing the concentrated cells while controlling the temperature. Similar to other types of samples, the frozen cells are cryopreserved in a liquid or gas-phase nitrogen freezer at -197°C. Similar to the method adopted by public cord blood banks for clinical use, the cryopreserved cells are defrosted, and the defrosted cells are transplanted into patients in cord blood stem cell transplantation.
Mononuclear cells consist mainly of lymphocytes and monocytes. However, CD34 positive cells are also in this fraction. Cord blood is layered on top of a reagent called Ficoll, centrifuged, and the obtained layer of mononuclear leukocytes is collected and cryopreserved. Mononuclear cells are easy to handle even after being defrosted, so these cells are widely used.
CD34 positive cells possess a typical marker of hematopoietic stem cells. After being purified using beads with antibodies, CD34 positive cells are cryopreserved. CD34 positive cells have attracted the interest of many researchers for conducting research on hematopoietic stem cell transplantation and blood cell separation, and as a source of induced pluripotent stem cells in the field of regenerative medicine.
|
Quality Control and Ethical Considerations |
|
Quality control of the cord blood is the most important duty of the Research Cord Blood Stem Cells Bank, because the quality of cord blood directly affects the results obtained in research using cord blood. Based on a medical interview of the mother, her family history, and her delivery record, the cord blood is judged to be normal or not when the baby is born. The cord blood is also tested for infectious diseases. The cell preparation methods have been developed according to the cell types. After the quality of the cord blood has been standardized, the samples are prepared.
Because these are human-derived cells, cord blood is strictly anonymized, and ethical practices are enforced at collection facilities, the Research Cord Blood Stem Cells Bank, and the RIKEN BRC. We ask all researchers who will use our samples to obtain ethical approval from their institution’s ethical review committee.
|
Toward Future Expansion of Use |
The results of a survey on the use of cord blood samples administered in fiscal year 2014 revealed that medical scientists (mainly physicians) used the samples the most, accounting for nearly 75% of the total users. The reason for this is probably that human-derived cells are primarily used in fields associated with clinical medicine or closely related to medical treatment. Therefore, the samples are used by a more specific group of scientists compared to the other various resources in the NBRP. We are expecting that the samples will soon be used more widely in the fields of basic research, in which it is difficult to obtain human cells.
|
|
|
|
From AmiGO 1.8 to 2.0
|

AmiGO is a web application developed by the Gene Ontology (GO) Consortium that can search, browse, and visualize ontologies. A stable release of AmiGO version 2.0 became available in March 2014 (※1), in which the user interface and the backend system has undergone significant changes from the previous version, 1.8 (※2). This article introduces some of the features and changes present in AmiGO version 2.0.
|

Firstly, we will discuss GO terms, annotations, and how to search for gene products.
1. Open the AmiGO2 web page (※3). You will see a large text field where you can enter queries and a [Search] button to be clicked to perform the query (Fig.1).
2. Select the target for the query from one of: "Ontology," "Genes and gene products," and "Annotations" (Fig.2).
3. In this example, "Annotations" of the gene or gene products relating to the query term "p53" are being shown (Fig.3).
In Fig. 3: ① shows the query term; ② is where you select the query criteria; ③ is where you can narrow your search result using filters; ④ is the page navigator; and ⑤is the list showing the query output. In the area indicated by ③, users can easily filter the query result, making it easy to obtain the desired result quickly.
In the example shown, adding "Homo sapiens" in the "Taxon" filter can narrow the search result from 6,761 hits to 1,672. |
|
Fig. 1. AmiGO 2’s simple search interface.
Fig. 2. Selecting target documents to narrow the search.
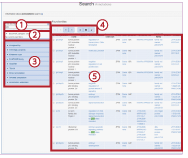
Fig. 3. Search results |

Next, I will briefly explain the backend system. The previous version, AmiGO 1.8, used SQL technology based on MySQL. However, provision of service while attempting to maintain an acceptable performance level using data caching and various other performance tuning techniques is becoming increasingly difficult. This is because of an over 50-fold increase in data volume, and because instances of complex queries having become more common.
In order to address this problem, AmiGO 2 has been re-architected to use GOlr as its backend system, and Perl and JavaScript as its front-end. GOlr is a search platform that uses Solr to index GO data, and it can be accessed using both Perl and JavaScript. This change has resulted in a marked improvement in search time; for example, a text search query that took 30 seconds using SQL now takes just 0.3 seconds. Additionally, the AmiGO 2 JavaScript API is also available (※4).
If you are interested in learning more about the technical details of the new version, please refer to the presentation material (※5) from the 13th Annual Bioinformatics Open Source Conference (BOSC 2012).
Why not take this opportunity to test the improved AmiGO?
(Manabu Kimura)
(Gaku Kimura)
|
|
|