|
 |
■ Bioresources information is available at the following URLs
|
 |
|
Introduction to Resource Center <No. 55> National BioResource Project Drosophila - Expansion of New Resources
Ryu Ueda (Professor, Genetic Resource Center, National Institute of Genetics)
 The National BioResource Project (NBRP) Drosophila consists of the National Institute of Genetics (NIG) and four associated facilities. The NIG has collected and delivered comprehensive mutant libraries (RNAi mutants) of Drosophila melanogaster. The RNAi mutant creation project launched in 2001 inserted a long-chain cDNA fragment (500 bp) into a vector (pUAST-R57) in the form of an inverted repeat sequence, which was considered effective, and integrated the vector into the chromosome using a P-element transposon. Until 2007, the NBRP Drosophila had continuously created vectors and established 12,741 fly stocks corresponding to 6,635 genes, into which the created vectors were integrated. The NBRP Drosophila has successively exhibited the fly stocks since fiscal year (FY) 2005, and had delivered approximately 130,000 fly stocks to foreign and domestic researchers until FY 2013. The National BioResource Project (NBRP) Drosophila consists of the National Institute of Genetics (NIG) and four associated facilities. The NIG has collected and delivered comprehensive mutant libraries (RNAi mutants) of Drosophila melanogaster. The RNAi mutant creation project launched in 2001 inserted a long-chain cDNA fragment (500 bp) into a vector (pUAST-R57) in the form of an inverted repeat sequence, which was considered effective, and integrated the vector into the chromosome using a P-element transposon. Until 2007, the NBRP Drosophila had continuously created vectors and established 12,741 fly stocks corresponding to 6,635 genes, into which the created vectors were integrated. The NBRP Drosophila has successively exhibited the fly stocks since fiscal year (FY) 2005, and had delivered approximately 130,000 fly stocks to foreign and domestic researchers until FY 2013.
|
 The number of protein genes in D. melanogaster is estimated to be 13,918 (FlyBase Release 6.02 in May 2014). To increase the cover ratio of RNAi mutants, the NBRP Drosophila began to create mutants in collaboration with the Transgenic RNAi Project (TRiP) at Harvard Medical School. TRiP has improved the efficiency of RNAi and developed various vectors. The number of protein genes in D. melanogaster is estimated to be 13,918 (FlyBase Release 6.02 in May 2014). To increase the cover ratio of RNAi mutants, the NBRP Drosophila began to create mutants in collaboration with the Transgenic RNAi Project (TRiP) at Harvard Medical School. TRiP has improved the efficiency of RNAi and developed various vectors.
The Valium20 vector announced in 2008 is a short hairpin (sh)-type RNAi vector using the skeleton of microRNA. Insulator sequences were inserted into the Valium20 vector to improve the expression-induction efficiency of shRNA using the GAL4-UAS system, resulting in the increased knockdown efficiency of a target gene (Fig. 1). A library of plasmids was available, in which a synthesized RNA oligonucleotide including 21-bp shRNA designed for exons of all protein genes, was inserted into the skeleton of the microRNA. However, only several thousands of plasmids were actually created. |
|
Fig. 1. Structure of the Valium20 vector
The upper drawing shows the layout of shRNA in the skeleton of the microRNA. The vector uses the vermilion + gene (orange) as a selective marker and possesses gypsy insulators (purple) on both sides. To control the expression level in the GAL4-UAS system, one of two 5xUASs (white) may be removed using the Cre/loxP method.
|
 In fact, this plasmid library was full of defectives, due to the new synthesis method. Subsequently, Assistant Professor Kondo from our laboratory, in cooperation with the NIG Advanced Genomics Center, began to sequence each clone on a plate. He sequenced approximately 70,000 clones and obtained 3,903 unique clones. Unlike conventional P-element transposons that had been used for gene transfer, the Valium20 vector used phiC31 recombinase∗1. Since only one type of DNA fragment was integrated into the attP site, which had already been embedded into the chromosome, a library, in which all useful clones had been mixed, was injected into a host egg, and all flies, whose eye colors had returned to those of wild flies due to the wild-type vermilion gene (the marker of the vector), were established as stocks. The genomic DNA of these flies was sequenced, and each stock was identified; consequently, mutant libraries could be efficiently constructed. In fact, this plasmid library was full of defectives, due to the new synthesis method. Subsequently, Assistant Professor Kondo from our laboratory, in cooperation with the NIG Advanced Genomics Center, began to sequence each clone on a plate. He sequenced approximately 70,000 clones and obtained 3,903 unique clones. Unlike conventional P-element transposons that had been used for gene transfer, the Valium20 vector used phiC31 recombinase∗1. Since only one type of DNA fragment was integrated into the attP site, which had already been embedded into the chromosome, a library, in which all useful clones had been mixed, was injected into a host egg, and all flies, whose eye colors had returned to those of wild flies due to the wild-type vermilion gene (the marker of the vector), were established as stocks. The genomic DNA of these flies was sequenced, and each stock was identified; consequently, mutant libraries could be efficiently constructed.
At present, the NIG has already collected 5,125 stocks corresponding to 4,382 genes, including stocks that were already established by TRiP. In addition, stocks established through this collaboration are shared with TRiP, and flies preserved by TRiP will be collected and exhibited by the Bloomington Drosophila Stock Center.
|
|
 To exhibit TRiP stocks, the 21-bp sequence was searched in the latest genome annotation data to confirm which mRNA was targeted in the target gene and its multiple transcriptional products. In summary, 8,860 genes were able to use the RNAi stocks. Out of the 8,860 genes, 2,157 genes were able to use both R57-RNAi and Valium-RNAi stocks (Fig. 2). At present, the NIG Center for Information Biology is creating a website for these genes, and will be open to the public soon. To exhibit TRiP stocks, the 21-bp sequence was searched in the latest genome annotation data to confirm which mRNA was targeted in the target gene and its multiple transcriptional products. In summary, 8,860 genes were able to use the RNAi stocks. Out of the 8,860 genes, 2,157 genes were able to use both R57-RNAi and Valium-RNAi stocks (Fig. 2). At present, the NIG Center for Information Biology is creating a website for these genes, and will be open to the public soon.
|
|
Fig. 2. Old and new cover ratios of the D. melanogaster genome in RNAi mutant stocks.
Out of the 13,918 genes in the genome, 12,741 R57-RNAi stocks target 6,635 genes, while 5,125 Valium-RNAi stocks target 4,382 genes. R57-RNAi and Valium-RNAi stocks target 8,860 genes in total.
|
|
 2013 was an epochal year for genome editing technologies. In particular, it was confirmed that CRISPR/Cas9 endonuclease could be used for various species. Various methods using this enzyme were examined for D. melanogaster. Assistant Professor Kondo found that mutation could be induced in F1 progenies at extremely high frequencies (60% on average) by crossbreeding a transgenic strain that expressed Cas9 in reproductive cells and a transgenic strain in which guideRNA had already been integrated (Fig. 3). After releasing information about the Cas9 stock and the vector for Drosophila transformation for the integration of the guideRNA used in this method online, Assistant Professor Kondo distributed 138 orders of 754 fly stocks and 115 orders of 247 vectors in 15 months. 2013 was an epochal year for genome editing technologies. In particular, it was confirmed that CRISPR/Cas9 endonuclease could be used for various species. Various methods using this enzyme were examined for D. melanogaster. Assistant Professor Kondo found that mutation could be induced in F1 progenies at extremely high frequencies (60% on average) by crossbreeding a transgenic strain that expressed Cas9 in reproductive cells and a transgenic strain in which guideRNA had already been integrated (Fig. 3). After releasing information about the Cas9 stock and the vector for Drosophila transformation for the integration of the guideRNA used in this method online, Assistant Professor Kondo distributed 138 orders of 754 fly stocks and 115 orders of 247 vectors in 15 months.
|
|
Fig. 3. Mutant creation using Cas9
When a fly in which Cas9 is expressed in reproductive cells by the nanos promoter and a fly that expresses guideRNA in all cells are crossbred (the upper part), the target gene is mutated in the reproductive cells of the next generation (the middle part), and, consequently, mutants appear at high frequencies in the F1 progenies (the bottom part). Since the white gene on the X chromosome is targeted in this figure, a phenotype directly appears in the males of the F1 progenies. |
 Our next tasks are to apply this genome editing technology to all genes to create a library of null mutants∗2 and to apply this technology to other Drosophila species. For the latter, we have been sequencing the genomes of 10 Drosophila species in the NBRP Genome Information Upgrading Program in this fiscal year in cooperation with the Drosophila research community. The information obtained will be released in the next fiscal year. Our next tasks are to apply this genome editing technology to all genes to create a library of null mutants∗2 and to apply this technology to other Drosophila species. For the latter, we have been sequencing the genomes of 10 Drosophila species in the NBRP Genome Information Upgrading Program in this fiscal year in cooperation with the Drosophila research community. The information obtained will be released in the next fiscal year.
|
| ∗1 |
An enzyme that catalyzes recombination occurring between attB donor plasmid, into which the objective gene has been integrated, and the attP site of the target genome. The enzyme is characteristic of linkage by a single copy (one piece), regardless of plasmid size and the site.
|
| ∗2 |
A mutant in which the function of the gene is entirely deleted. |
|
|
Be aware of Facebook’s privacy settings |

Facebook is a popular social networking service (SNS) used worldwide, and there must be many users among our readers as well. Facebook can be used not only by individual users for sharing information but also by companies and organizations for promotional activities targeting specific users. In the summer of 2014, NBRP Tomato launched its own Facebook page to enable real-time dissemination of information
There are many settings to consider when using Facebook, and it is easy to overlook a number of its privacy settings. In this edition, I will introduce a few of the lesser-known privacy settings.

1. Most users would already be restricting who can view their posts to prevent unnecessary disclosure of information. Users can configure the privacy setting of individual posts, a feature that should encourage people to think about the intended audience of various contents before submitting each post.
|
2. Did you know that when you search your own name using a search engine, such as Google, your Facebook pages could appear in the list of results? You can limit who can view your posts. Furthermore, you can even prevent your Facebook page from appearing in the search result in the first place.
Select [Settings] > [Privacy] > [Do you want other search engines to link to your timeline?] > [Edit] > [No] (Fig. 1).
|
|
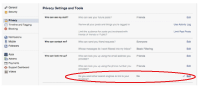
Fig. 1. Privacy setting to hide your links from search engines.
|
3. There are a number of features currently available only in the US, but these will become available in Japan after an update in the user policy planned for 2015. One such setting is the ability to control the display of ads in a user’s timeline based on the same users’ browsing history.
For example, if you were searching for the word “chicken” on the web, ads relating to “chicken” might be displayed in your timeline. There would be users who would prefer not to have the system utilize their browsing history without explicit authorization. The opt-out setting for preventing the use of browsing history for ads in the US must be done from a website that is external to Facebook. The opt-out procedure is described below.

Digital Advertising Alliance (US)
http://www.aboutads.info/choices/
- From the above URL, click on the tab titled [Companies Customizing Ads For Your Browser](Fig. 2-A).
- Tick the checkbox next to "Facebook" (Fig. 2-B).
- Click on [Submit your choices] (Fig. 2-C).
This process completes the opt-out setting for Facebook.
You can also opt out of services other than Facebook (e.g., Google) by selecting the respective checkboxes (Fig. 3).
|
| |
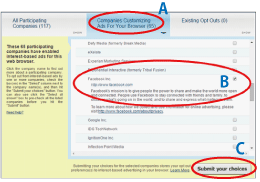
Fig. 2. Opt-out setting.
| |
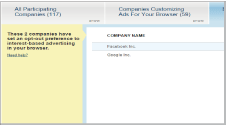
Fig. 3. Opt-out setting for other services |

Finally, please be wary of the fact that once you have submitted a post, you are disclosing information to Facebook, an external organization. Even if you can choose who can view your posts, Facebook still has your information. It is advisable that you re-think how you use SNSs and review your privacy settings.
(Mika Nagahora, Genetic Resource Center, Database Division) |
|
|
BRW (BioResource World)
|
・Number of registered resources : 6,372,945
(As of December,2014) |
| DB名: |
BRW (BioResource World) |
| URL: |
http://resourcedb.nbrp.jp/ |
| Language : |
Japanese, English |
| Contents : |
・ |
Integrated search for resources in each site of the National BioResearch Project (NBRP) |
| Features : |
・ |
All resources in each site of the NBRP may be searched and browsed. |
| |
・ |
BLAST search may be performed for information about sequences in the clone lists in each site of the NBRP. |
|
・ |
In addition to a keyword search, resources may be searched from Gene Ontology (GO), Taxonomy, and Disease Ontology (DO) |
| |
・ |
Resources may be searched from the NBRP Reference. |
| Cooperative DB : Each site of the NBRP, RRC |
| DB construction group : NBRP Information |
| Management organization : Genetic Resource Center, NIG |
| Year of first DB publication : 2004 Year of last DB update : 2014 |
|
Comment from a developer : BRW is an integrated search site for resources in each site of the NBRP. BRW was opened to the public in December 2004, so 10 years have passed since its inauguration. When BRW was first opened to the public, only strain resources could be searched. In 2006, clones could also be searched. In 2007, 2009, 2011, 2012, and 2013, BLAST search, GO search, Taxonomy search, Reference search, and DO search became available, respectively. At present, resources may be searched from various aspects. In parallel, search functions have been enhanced. At the beginning, only the SQL search function was available. By adopting the Oracle search function, Shunsaku, and Memcached, resources may be searched more accurately and more quickly than previously. In the future, we will not only increase the accuracy and speed, but also enable users to narrow down search results by adopting the Solr search engine. The test version of the Solr search engine is available at resourcedb.nbrp.jp/v2/.
BRW has been improved along with the advancement of the NBRP. Please feel free to use BRW, and do not hesitate to express your comments, questions, or opinions.
| |
|