|
 |
■ Bioresources information is available at the following URLs
|
 |
|
Introduction to Resource Center <No. 54> Research on Cellular Slime Molds and Changes in Resource Project
Hideko Urushihara ,Professor, Faculty of Life and Environmental Sciences, University of Tsukuba
|
Cellular slime molds, which are used as model organisms in the field of medicobiology, are amoeba-like eukaryotic microorganisms that live in soil and proliferate by preying on microorganisms such as bacteria. The notable characteristic of cellular slime molds is that under starvation conditions, after the surrounding feed has been consumed, these organisms aggregate to form a multicellular fruiting body referred to as a "sorocarp." Because role sharing between spores and the stalk cells that support a spore clump has been observed, cellular slime molds are called "social amoebae."
The resource project for cellular slime molds was launched in August 2007, at the beginning of the second term of the National BioResource Project (NBRP). Therefore, as of November 2014, seven years have passed. The outline of this resource project was introduced in a previous newsletter (2008). Subsequently, various circumstances have changed surrounding the NBRP Cellular Slime Molds. In this article, I will introduce several topics regarding these changes as well as recent advances in the research on cellular slime molds. |
Further Diversification of Research Using Cellular Slime Molds |
|
Culturing of cellular slime molds is cheap and easy, and multiple biochemical and molecular biological methods can be applied to study these organisms. As a result, cellular slime molds have been used in various fields of fundamental and applied sciences. Recently, excellent results have been achieved using advanced technologies, such as signal transduction, in which markedly advanced bioimaging and single-molecule measurements are combined, and single-cell transcription analysis. Interesting findings have also been reported by directly observing cellular slime molds. For example, a phenomenon termed “farming” was discovered in which spores do not consume bacteria, but carried a part of bacteria to a new place, and proliferated the carried bacteria in the new place before germinating. This phenomenon is an extremely interesting survival strategy adopted by cellular slime molds, which can be used to study interactions among bacteria, cellular slime molds, and plants, and is applicable to agricultural sciences. |

Fig. 1. Waves of cellular slime molds, referred to as "biological soliton" waves.
Waves indicated by the yellow and black arrows approach from left and right, respectively, collide, and pass through each other
(these photographs are provided by Dr. Hidekazu Kuwayama) |
At appropriate cell densities, certain non-chemotactic Dictyostelium discoideum mutants that are unable to form normal aggregates have been observed to move collectively in one direction, like a wave. When two waves collide, however, they pass through each other. This phenomenon is similar to the characteristics of soliton waves in physics (Fig. 1). This "biological soliton" has therefore attracted the interest of researchers who examine the boundary regions between biology and physics. A new study on cellular slime molds, which is entirely different from previous studies, is expected to be developed.
Similar to Escherichia coli, which is used as a host for genetic cloning, some researchers use D. discoideum as an operational platform and an experimental system for evaluation. These are new studies conducted by new users. For example, D. discoideum has been used to examine the intracellular behavior, stability, and color development of a new fluorescent protein in its development process by expressing it in D. discoideum cells. Furthermore, that D. discoideum cells react sensitively to cyclic adenosine monophosphate during the aggregation stage has been exploited to evaluate photoresponsive proteins. The NBRP Cellular Slime Molds, which aims to expand the scope of research using cellular slime molds, has provided a training course, primarily for new users, every year since 2011, and we are beginning to see the results. |
Ironic Relationship between Technological Progress and Achievements of the NBRP Cellular Slime Molds |
|
Owing to the diffusion of next-generation sequencers, the NBRP Cellular Slime Molds is currently making the genome sequences of four species of cellular slime molds, in addition to the genome sequence of the D. discoideum type species, available to the public. Advancements in the development of genomic and molecular biological technologies should be welcomed. Ironically, however, the number of gene clones distributed by the resource project has decreased. A requirement for users to pay the necessary cost for distribution of resources, which began in 2010, has accelerated this tendency. The number of resources requested from overseas, which accounted for nearly all of the requests prior to 2010, has decreased sharply during this period. In contrast, the Dicty Stock Center, which is a central repository for D. discoideum strains in the United States that began distribution services five years earlier than the NBRP Cellular Slime Molds, distributes resources at no charge. However, under such circumstances, we are forced to ask what the goals of the resource project should be. At present, the NBRP Cellular Slime Molds is stepping up efforts to collect various expression vectors (Fig. 2) as further resources to be requested in the future, and wild strains (Fig. 3), which could expand the scope of research of cellular slime molds and which are considered as treasure troves of useful substances. |
| |
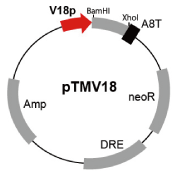
Fig. 2. Vector map of pTMV18 (G90126). This vector contains the V18 promoter for induction of gene expression during the proliferation stage. Numerous cellular slime mold expression vectors are suitable for gene expression during the developmental stage. |
|
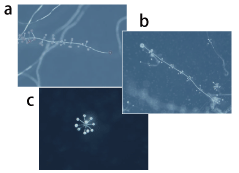
Fig. 3. Wild strains of cellular slime molds.
a: Polysphondylium tenuissimum, b: Polysphondylium violaceum, c: Dictyostelium polycephalum
|
The Great East Japan Earthquake and the Development of a New System |
Since its establishment, the most detrimental event of the NBRP Cellular Slime Molds was the Great East Japan Earthquake that occurred on March 11, 2011.
Because both the core center (Faculty of Life and Environmental Sciences, University of Tsukuba) and the sub-center (Research Institute for Cell Engineering, National Institute of Advanced Industrial Science and Technology) were located in Tsukuba City when the earthquake occurred, although the core and the sub-center had mutually created backups, we lost dozens of strains that had not been preserved in liquid nitrogen and whose freeze-dried spores had not been stocked.
With the effort made by the sub-center, we managed to re-collect and reproduce almost all of the lost strains.
However, a great deal of time and manpower were spent. We learned a lesson from this accident and, as a result, have thoroughly revised the system of preserving resources. Consequently, we have established a new sub-center (RIKEN Quantitative Biology Center) at Osaka University.
In the newly established center, we have preserved resources for backup and updated stocks, and have installed a generator, the cost of which was paid by the university. Furthermore, we aim to use all of the three methods for preserving cellular slime mold strains (using a freezer, using liquid nitrogen, and in the form of freeze-dried spores), if possible.
We have improved the preservation conditions for each strain, completed the database for backup resources, and prepared a resource management manual.
Three and a half years have passed since the Great East Japan Earthquake, and we barely remember the miserable situation at that time.
Even so, everyone involved in the resource project will make every effort to preserve the resources although we might not consciously remind ourselves daily.
|
|
|
What you can do with Bio-Linux |

In the previous edition, we described the steps required to create a virtual environment to run Bio-Linux. This edition introduces some genome sequence analysis tools that are pre-installed with Bio-Linux.

|
1. Pre-installed commands
You can perform homology searches, create phylogenetic tree diagrams, and conduct many other tasks. Over 250 tools are available in the Bio-Linux standard installation. Please refer to the list of tools in the “Bio-Linux Documentation,” as mentioned later in this article.
2. Launching the terminal window
The tools introduced in this article are all executed via the command line inside a terminal window. The terminal window can be opened by clicking on the icon on the top left-hand corner, in the application bar (Fig 1-①).
3. Manipulating sequences
Commands from the EMBOSS package can be used to manipulate sequences, such as revseq (reverses and complements a sequence), extractseq (extracts regions from a sequence), and maskseq (writes a mask version of a specified region). Please refer to the "Bio-Linux Documentation" mentioned later in this article for further information about how to use these tools.
4. Exporting BLAST as a tab-delimited file
This popular tool is used to search for homologous sequences. The BLAST output can be exported into various file formats from the command line. The following examples use nucleic acid sequences.
① Formatting the database
Prepare a FASTA file of the sequence (database) that you would like to search against, type the following command into the terminal window and press Enter.
makeblastdb –in FASTA {filename} –out {database name} –dbtype nucl
② Executing blastn
Prepare a FASTA file of the query containing the sequence that you would like to search for. Type the following command into the terminal window and press Enter.
blastn –db {database name} –query {query file} –outfmt 7 –out {output file}
The search result will be output to a file. You can open the tab-delimited output file using spreadsheet software such as LibreOffice Calc ((Fig. 1-②, a program that is equivalent to Microsoft Excel, which is installed with Bio-Linux) for better readability (Fig. 2). |

Fig. 1. The application bar |
|
5. Creating phylogenetic tree diagrams from alignments with CLUSTALW
CLUSTALW is a multiple sequence alignment tool with a very long history.
① Execute clustalw
Prepare a Multi-FASTA format file containing the sequences that you would like to align. Enter the following command into the terminal window and press Enter.
clustalw –INFILE={input file} –OUTFILE={output file}
The results are written to an output file (Fig. 3).
② Generate the file for creating a phylogenetic tree diagram
Type the following command into the terminal window and press Enter.
clustalw –INFILE={input file} –tree
A similar message to the following will be displayed after the command has completed.
Phylogenetic tree file created: [test.ph]
③ Generate the phylogenetic tree
Execute the following command from the terminal window.
treeview test.ph (this is the file generated in step 5-② above)
The tree diagram will be displayed in a new window (Fig. 4).
|
 Fig. 2. Output from blastn

Fig. 3. Output from clustalw
Fig. 4. Output from treeview |

Many other tools are packaged within Bio-Linux. A list of these tools and explanations of how to use them can be viewed by double-clicking on "Bio-Linux Documentation" on the desktop, and then double-clicking on "Bioinformatics Docs." There are many useful tools to assist your research, so it is worth taking a look.
(Shunnsuke Maeda) |
|
|
JMSR (Japan Mouse/Rat Strain Resources Database)
|
| |

・ Number of strains :9,301
(As of November, 2014) |
| DBname : |
JMSR (Japan Mouse/Rat Strain Resources Database) |
| URL : |
http://www.shigen.nig.ac.jp/mouse/jmsr/ |
| Language : |
Japanese, English |
| Original contents: |
| |
・ |
A portal site for mouse/rat strain resources available in Japan. |
| |
・ |
The database contains no original contents. |
| Features : |
・ |
Nearly all mouse/rat strain resources available in Japan can be searched and ordered via links to the distributing organizations. |
| |
・ |
Strain resources can be searched by strain types, types of stocks, and animal models for human diseases. |
| |
・ |
Data are appropriately updated in cooperation with stock centers and databases. |
| Cooperative DB : |
| |
NIG (National Institute of Genetics), CARD (Kumamoto University), KYOTO RAT (Kyoto University), BRC (Riken BioResource Center), CRLJ (Charles River Laboratories Japan, Inc.), NIBIO, JCRB (National Institute of Biomedical Innovation), JCL (CLEA Japan, Inc.), TG Resource Bank (Trans Genic Inc.), SLC (Japan SLC, Inc.), and OBS (Oriental Bioservice, Inc.) |
| DB construction group : |
| |
Mouse Genetic Resources Subcommittee,
Genetic Resource Center, National Institute of Genetics |
| Management organization : Genetic Resource Center, NIG |
| Year of first DB publication : 2001 Year of last DB update : 2014 |
|
Comment from a developer:
Among bioresources for research purposes, there is a particularly large number of researchers using the mouse/rat DB.
In Japan, several organizations distribute mouse/rat strain resources.
The JMSR was established after the second meeting of the Mouse Genetic Resources Subcommittee, and opened to public in 2001 as a website which enables batch searching for mouse/rat strain resources.
The JMSR can be said to be the Japanese version of the International Mouse Strain Resource (IMSR).
When first opened to the public, there were only four organizations participating in the JMSR.
At present, however, 10 organizations participate in the JMSR.
Namely, this database covers all organizations that distribute mouse/rat strain resources.
Data have been updated on more than 200 occasions, and the number of strains listed is approximately 10,000.
The JMSR has been stably managed for nearly 15 years.
At present, we have a plan in place to update the website and to expand the search functions.
When you search for mouse/rat strain resources, please visit the JMSR by all means!
| |
|