|
 |
■ Bioresources information is available at the following URLs
|
 |
|
Research and Bioresources <No. 18> The Evolution of Sex-Determining Genes from Diversity in Medaka
Yusuke Takehana (Assistant Professor, Laboratory of Bioresources, National Institute for Basic Biology)
|
Approximately 30 species of medaka are widely distributed in Asia, ranging from India to Japan, including the Japanese medaka (killifish, Oryzias latipes). The National Bioresource Project (NBRP) Medaka has preserved 13 species and 20 strains out of these 30 existing species. We previously demonstrated that the sex chromosomes of Oryzias fishes are extremely diversified. Here, we elucidate part of the diversification mechanism of sex chromosomes by identifying sex-determining genes in the Indian medaka (O. dancena), which is widely distributed from East India to the Malay Peninsula. Here, I introduce the content of our research. |
Sex Determination and Sex-Determining Genes in Vertebrates |
|
In most vertebrates, males and females are determined by sex chromosomes. For example, in mammals, if individuals possess XX and XY sex chromosomes, they become males and females, respectively. The male specific Y chromosome contains the Sry gene, which determines sex, with differentiation into a male being initiated by the functioning of this gene. However, the Sry gene does not exist in other vertebrates, with the chromosome that functions as the sex chromosome differing across taxa. Sex determination is highly diversified in fish species. The sex chromosomes of Oryzias fishes are extremely diverse. A sex-determining gene, Dmy, was identified in the Japanese medaka, which was the second case in vertebrates. However, the Dmy gene only exists in two medaka species, with other related species possessing different sex chromosomes (Fig. 1).
This fact indicates that sex chromosomes and sex-determining genes have changed many times within a short period. Thus, we aimed to elucidate the change in the sex-determining mechanism at the molecular level. Specifically, we searched for the sex-determining gene in the Indian medaka (O. dancena), which did not possess the Dmy gene. |

Fig. 1. Phyletic relationships and the sex-determining mechanism in Oryzias fishes.
Sex chromosomes in Oryzias fishes differ according to species. The Dmy gene found in Oryzias latipes does not exist in other related species, except for O. curvinotus.
|
Identification of the Sex-Determining Gene in the Indian medaka |
|
Previous studies have revealed that the sex determination of the Indian medaka is the XY type, and that its sex chromosome is homologous with an autosome (the 10th chromosome). We used positional cloning to identify the sex-determining gene in the Indian medaka. First, we created a detailed genetic linkage map near the sex-determining locus. We also created a physical map near the sex-determining locus by using a bacterial artificial chromosome (BAC) genomic library of the Indian medaka, from which we determined the entire base sequence of this region. As a result, we were able to narrow down the sex-determining region (SD region) to 140 kb on the X chromosome and 310 kb on the Y chromosome (Fig. 2).
The SD region is filled with repeated sequences including transposons, with no known gene that encode proteins being found in this region. However, the Sox3 gene was found in a region adjacent to the SD region. When part of the Y-chromosome sequence, including the Sox3 gene and the SD region, was experimentally introduced to an XX individual, the sex of the individual changed to male. When the Sox3 gene on the Y chromosome was knocked out using genome editing techniques in an XY individual, the sex of the individual changed to female. These experiments indicate that the Sox3 gene is a sex-determining gene in the Indian medaka, and that the expression control region (enhancer) of the Sox3 gene exists in the SD region. In other words, the acquisition of the enhancer may generate a new Y chromosome. |
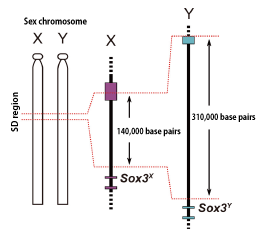
Figure 2. Sex chromosomes (X and Y) and the SD region in the Indian medaka
The SD region that is involved in sex determination consists of 140,000 base pairs on the X chromosome and 310,000 base pairs on the Y chromosome. The Sox3 gene exists near the SD region. The Sox3 gene exists on both the X (Sox3X) and Y (Sox3Y) chromosomes, with both chromosomes having the same amino acid sequence.
|
Sox3 Gene, an Ancestor of the Sry Gene |
The Sox3 gene is highly homogeneous with Sry, a sex-determining gene in mammals, and it encodes a transcription factor. Both genes (Sox3 and Sry) may have an ancestral allelic relationship, because both genes exist on the same locus. However, since the Sry gene is of extremely old origin, its past allelic relationship is unknown. In addition, the relationship between the Sox3 gene and sex determination had not been reported in vertebrates other than mammals. Our work may directly prove that a new sex-determining gene could be generated by functional differentiation between Sox3 alleles, showing that the same Sox3 gene has independently evolved as a sex-determining gene in mammals and in Oryzias fishes. |
Diversification Mechanism of Sex-Determining Genes |
|
In Oryzias fishes, two male-determining genes have been identified other than Sox3 in the Indian medaka. One is the Dmy gene that is found on the Y chromosome in the Japanese medaka (O. latipes) and the other is the Gsdf gene that is found on the Y chromosome in the Luzon medaka (O. luzonensis) (Fig. 3)
Although the Dmy gene only exists in two medaka species, the Gsdf gene is expressed in the testis of many fish species. In the Japanese medaka, the Gsdf gene is thought to be downstream of the Dmy gene. Here, we revealed that the Gsdf gene is also highly expressed in the gonad of the Indian medaka, and that its expression is dependent on the expression of the Sox3 gene on the Y chromosome. These results indicate that the sex-determining cascade below the Gsdf gene has been interspecifically preserved, and that a new sex-determining gene may have been generated through the mutation of the Gsdf gene or the acquisition of transcriptional regulatory elements for the Gsdf gene (Sox3 and Dmy). |
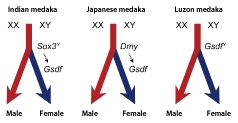
Fig. 3. Diversification of sex-determining genes in Oryzias fishes.
Three species from the genus Oryzias (i.e., the Indian medaka, the Japanese medaka, and the Luzon medaka) possess different male-determining genes (Sox3Y, Dmy, and GsdfY). However, the Gsdf gene is highly expressed in XY individuals of all three species. In the Indian medaka, the Sox3Y gene is assumed to control the expression of the Gsdf gene (which was located below the Sox3Y gene), resulting in the generation of a new sex-determination mechanism.
|
Following the discovery of the sex-determining gene in the Indian medaka, the diversification mechanism of a sex-determining gene and the preservative property of a downstream gene have been elucidated. However, many questions remain, such as whether the Gsdf gene is a common master gene in sex determination and how did the alternation between the XY type and ZW type occur. In the future, we intend to elucidate both the diversity and generality in the sex-determining mechanism by obtaining new information about other species in the genus Oryzias.
|
| |
Refernce |
| |
Takehana et al., Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nature Communications 5:4157 (2014)
|
|
|
Construction of Linux Virtual Environment in Windows |

Some time has passed since the use of next-generation sequencers became commonplace. Nevertheless, having a Linux environment is still a requisite for analyzing sequence data, because analysis tools often cannot be installed in a Windows environment. In this article, I shall introduce a method for installing a Linux virtual machine on Windows using VMware Player, a tool that is free for non-commercial use.
|
Installing VMware
1. Download the "VMware Player for Windows" installer from the download page∗1 (Fig.1).
2. Double-click on the installer to launch it once it has downloaded, and follow the instructions to complete the installation.
|
|

Fig. 1. Downloading VMware |
Obtaining a Linux distribution
While there are several Linux distributions available, we will use Bio-Linux 8, a distribution based on Ubuntu 14.04 that comes pre-installed with several useful analysis tools. Download the ISO file from the Bio-Linux download page∗2 (Fig.2).
Installing Bio-Linux on VMware
1. Launch VMware Player and click on [Create a New Virtual Machine].
2. Select "Installer disc image file (iso)," click on [Browse], and select the ISO file downloaded during the previous step. Then, click [Next].
3.Select "Linux" as the guest OS and "Ubuntu 64 bit" as the version. Proceed with the installation wizard by choosing other settings pertaining to your computer's environment.
4. After the wizard has completed the installation, the new virtual machine will be displayed (Fig.3). Select the virtual machine, and click on [Play Virtual Machine] to launch it.
5. In the screen that is displayed after launching the virtual machine, click on [Install Bio-Linux] to begin installing Bio-Linux (Fig.4).
6. Follow the instructions from the installation wizard to complete the installation.
∗ Because Bio-Linux is developed in English, make sure to select English as the language during the installation.
What you can do with Bio-Linux
Bio-Linux comes pre-installed with several tools. Double-click on "Bio-Linux Documentation" located on the desktop, and double-click on "Bioinformatics Docs" to view detailed explanations for the available tools (Fig.5).
|
|
Fig. 2. Downloading Bio-Linux
Fig. 3. VMware startup screen
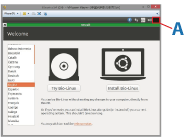 Fig. 4. Bio-Linux installation screen
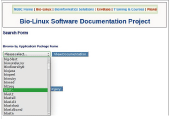
Fig. 5. Documentation for Bio-Linux tools, etc. |
Starting up and shutting down the virtual machine
In order to shut down the virtual machine, click on the cog icon in the top right-hand corner of the Linux desktop (Fig.4- A) and select [Shut Down…]. In order to start the virtual machine, click on the created virtual machine in the VMware Player window (Fig.3) and click on [Play Virtual Machine].
Configuring the virtual machine
You can configure various settings, such as those related to the memory and processor, by selecting the virtual machine on the VMware Player screen and clicking [Edit virtual machine settings].
In this edition, I have explained how to install Bio-Linux. In the next edition, I will introduce the steps required for performing analyses using some of the tools that come preinstalled with Bio-Linux.
|
(Shunsuke Maeda) |
|
RRC (Research Resource Circulation)
|
| |
・Number of registered papers: 14,801
・Number of registered patents: 862
(As of October 2014)
|
| DB name : |
RRC(Research Resource Circulation) |
| URL : |
http://rrc.nbrp.jp/ |
| Lungage : |
Japanese, English |
| Original contents : |
・ |
Information about research papers and patents obtained using the National Bioresource Project (NBRP) resources |
| Features : ・ |
Information about a paper can be registered. |
・ |
A research paper can be registered only with PubMed ID. |
・ |
From a research paper, information about the related resource can be accessed. |
・ |
RRC has been registered in PubMed LinkOut. |
| Cooperative DB :
Each site of the NBRP, NCBI PubMed |
| DB construction group : NBRP Information |
| Management organization : Genetic Resource Center, NIG |
| Year of first DB publication : 2007 Year of last DB update : 2014 |
|
Comment from a developer:: RRC is a database and a registration site for accessing information about research papers and patents obtained using the NBRP resources. In 2013, the RRC became a provider of the PubMed LinkOut, and a resource order page can be accessed from PubMed Abstract through the RRC. The NBRP is in the midst of developing a function whereby information about a research paper registered in the RRC is automatically added to the detailed page of each resource. Thus, a virtuous cycle of resources and research activities can be realized.
When a research paper or a patent obtained using the NBRP resources is published, please register it with the RRC.
| |
|