|
 |
■ Bioresources information is available at the following URLs
|
 |
|
CIMMYT Wheat Breeding and Japan's Contribution to World Wheat Production
Masahiro Kishii, Head of Global Wheat Program
(International Maize and Wheat Improvement Center)
 The International Maize and Wheat Improvement Center (CIMMYT; located in Mexico; Photo 1) is an international breeding center aiming to improve living standards of poor farmers in the world by providing better wheat and maize varieties. One of the achievements of the institute is a pioneer work on the development of semi-dwarf wheat varieties headed by Dr. Norman Borlaug that led to the Green Evolution. Since then, many of the wheat varieties produced in CIMMYT have been not only directly released to developing countries where their own breeding capacity has not been sufficient but also utilized as breeding materials in many industrialized counties. The International Maize and Wheat Improvement Center (CIMMYT; located in Mexico; Photo 1) is an international breeding center aiming to improve living standards of poor farmers in the world by providing better wheat and maize varieties. One of the achievements of the institute is a pioneer work on the development of semi-dwarf wheat varieties headed by Dr. Norman Borlaug that led to the Green Evolution. Since then, many of the wheat varieties produced in CIMMYT have been not only directly released to developing countries where their own breeding capacity has not been sufficient but also utilized as breeding materials in many industrialized counties.
 As predicted by a report that the world population will increase to 10.9-16.6 billion by the end of this century (UN DESA 2013;http://www.un.org/en/development/desa/index.html), it is necessary to keep releasing higher yield wheat varieties to satisfy increasing food demand. But in recent years it has become more difficult to catch up this mandate because of global climate change that would cause many abiotic stresses such as drought and heat. As predicted by a report that the world population will increase to 10.9-16.6 billion by the end of this century (UN DESA 2013;http://www.un.org/en/development/desa/index.html), it is necessary to keep releasing higher yield wheat varieties to satisfy increasing food demand. But in recent years it has become more difficult to catch up this mandate because of global climate change that would cause many abiotic stresses such as drought and heat.
 To overcome these difficulties, CIMMYT wheat-breeding program has been utilizing any kind of key methodologies that would be able to facilitate for releasing higher yield and stress resistance varieties, including molecular technique, bioinformatics (such as genomic selection), transgenic, physiological approaches, F1 hybrid wheat, improved agronomical practices and, of course, use of genetic resources (landraces, wheat ancestor and alien species). To overcome these difficulties, CIMMYT wheat-breeding program has been utilizing any kind of key methodologies that would be able to facilitate for releasing higher yield and stress resistance varieties, including molecular technique, bioinformatics (such as genomic selection), transgenic, physiological approaches, F1 hybrid wheat, improved agronomical practices and, of course, use of genetic resources (landraces, wheat ancestor and alien species).
|
| |

Photo 1: CIMMYT headquarter in Mexico |

Photo: Dr. Kishii in front of the Genebank Building
|
|
 It is not known by many Japanese people that Japan has been hugely helping the works of CIMMYT by providing infrastructures including its Genebank building (Photo 2). Much less people are aware of the more important fact that hundreds of accessions of wheat ancestor species (namely Aegilops tauschii) originally coming from NBRP-Wheat (or Kyoto University prior to the start of NBRP project) have been used to produce new synthetic bread wheats (man-made new bread wheat by crossing durum and Aegilops tauschii; Photo 3) for improvement of drought and other stresses (Photo 4). It is certain that many of additional genetic resources in NBRP-Wheat will contribute to world wheat production in the future. It is not known by many Japanese people that Japan has been hugely helping the works of CIMMYT by providing infrastructures including its Genebank building (Photo 2). Much less people are aware of the more important fact that hundreds of accessions of wheat ancestor species (namely Aegilops tauschii) originally coming from NBRP-Wheat (or Kyoto University prior to the start of NBRP project) have been used to produce new synthetic bread wheats (man-made new bread wheat by crossing durum and Aegilops tauschii; Photo 3) for improvement of drought and other stresses (Photo 4). It is certain that many of additional genetic resources in NBRP-Wheat will contribute to world wheat production in the future.
|

Photo 2: The front of the Genebank building in CIMMYT |

Photo 3: New synthetic wheat (right; green color as resistance for rust disease) and non-resistant wheat variety (left)
| 
Photo 4: Drought tolerance test in Obregon field.
The right side is for drought condition, and the left is for control with irrigation.
|
"This article is written in English by author."
|
|
Easy and Free Mapping of NGS Data to Genomic Data ∼Part 1∼ |
Advances in next-generation sequencing (NGS) methods and their wide availability have led to a greater number of sequence databases, making it easier to acquire NGS data in recent years. I will introduce techniques for mapping NGS data to genome sequences and viewing the results using a genome viewer. The article is split into two parts, with Part 1 being presented this month.
| ※ |
Although examples in this article use Windows (validated in Windows 7 and 8), the software being introduced also supports Mac and Linux operating systems. |
|
Mapping sequence data to genomes
A tool called Bowtie2 will be used for mapping sequence data.
| |
In the example below, a folder called "test" is created directly under the C:\ drive. Downloaded zip files as well as unzipped files are stored in this folder. Please use an appropriate tool for extracting the contents of the downloaded archives. |
|
 Obtaining the software Obtaining the software
You can download Bowtie2 from the following URL:
http://sourceforge.net/projects/bowtie-bio/files/bowtie2/2.1.0/
Download bowtie2-2.1.0-mingw-winXX.zip(XX=64 for 64-bit OS or 32 for 32-bit OS) and extract the contents to a folder.
Open [Control Panel → System → Advanced System Settings] and click on [Environmental Variables]. Click on the "Path" variable within [System variables] and click on [Edit]. Add a semicolon (";") at the end of the input field and add the path to the unzipped content (Fig. 1). Please do not make changes to any of the existing path information.
| 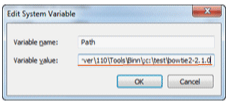
Fig. 1: Editing system variables |
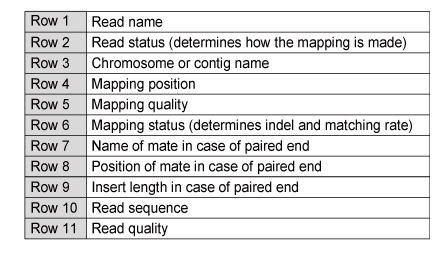
The generated SAM file is very large: please use a viewer that can handle large files. The standard fields in the SAM file are given below in Table 2.
Table 2. The contents of a SAM file
In Part 2 of this article, I will introduce Integrative Genome Viewer (IGV), which can be used to visualize the mapping results contained in the generated SAM file.
(Shunsuke Maeda, Genetic Resource Center) |
|
|
|
National BioResource Project "Zebrafish"
|
▪ Number of strains: 357
(as of October 2013) |
|
| DB name: |
NBRP Zebrafish |
| URL: |
http://www.shigen.nig.ac.jp/zebra/ |
| Language: |
Japanese, English |
| Contents: |
▪ |
Zebrafish strain resources for research
(wild, mutant, and transgenic strains) |
| ▪ |
Useful information on zebrafish
(zebrafish-related sites, protocols, seminars, and laboratories) |
| ▪ |
Newsletter |
| Features: |
You can order resources from this database.
Direct link with the Zebrafish Model Organism Database (ZFIN) enables you to obtain zebrafish-related information. |
| Cooperative DB: ZFIN, zTRAP |
| DB construction group: NBRP Zebrafish, NBRP Information |
| Management organization: Genetic Resource Center, NIG |
| Year of first DB publication: 2004 Year of last DB update: 2013 |
|
Comment from a developer : Although the NBRP Zebrafish is a small and simple database in terms of the amount of data and information items, the database can provide sufficient information to users, in cooperation with ZFIN. The database is managed by the Center for Genetic Resource Information, National Institute of Genetics. Since information on zebrafish is managed by NBRP Zebrafish, the latest information can be provided. In the future, we will add research papers from users of our resources to the database. Please feel free to use the database, and do not hesitate to send us your comments, questions, or opinions via Contact Us on the left-hand side menu.
|
|
|