|
 |
■ Bioresources information is available at the following URLs
|
 |
|
Introduction to Resource Center <No.39> BioResource Center for E. coli and B. subtilis, model prokaryote organisms
Hironori Niki (Professor, Genetic Strains Research Center, National Institute of Genetics)
Tsutomu Katayama (Professor, Graduate School of Pharmaceutical Sciences, Kyushu University)
Ten years have passed since the inauguration of the National BioResource Project (NBRP). At the beginning of the project, our primary bioresources were mutant Escherichia coli strains at the Genetic Stock Research Center, National Institute of Genetics (NIG). Since then, we have obtained resources in the post-genome era that were developed using genomic information. These resources include a collection of E. coli strains that contains a mutant for each of its genes. In addition, we have collected knockout Bacillus subtilis strains and are now send more than 200,000 samples to researchers across the globe every year (Fig. 1). NIG has been primarily responsible for preserving and distributing the resources, while Kyushu University has been preserving a set of the resources as a protective backup.
|
Genome-wide bioresources of E. coli and B. subtilis |
| Development of deletion mutants for all E. coli and B. subtilis genes began immediately after completion of the sequencing of their genomes. As estimated from the gene sequences, each bacteria has over 4,000 genes. By deleting the genes one by one, we have been able to determine which genes are essential for growth.
There are 2 collections of E. coli knockout strains. In one collection, each gene region was replaced by drug-resistance genes (KEIO collection). In the other collection, transposons were inserted into each gene. Obviously, knockout strains of essential genes cannot be developed by replacing them with drug-resistance genes. In order to resolve this issue, transposon insertion–deletion mutant collections have been developed by partially duplicating a chromosome region and inserting transposons into a gene in the duplicated region. The duplicated region can then be eliminated by recombinant reactions. This method is useful in that the recombinant strains can be selected by culturing the strains at a temperature of 37°C or more, and the effect of the gene deletion can be evaluated.
B. subtilis knockout strains have been developed by artificially inserting drug-resistance genes into each target gene by using a recombination technique. However, only about 2,000 mutant strains are currently available. Mutant strains for about 1,900 genes are not yet included in the collection. Completion of this collection is one of the important future tasks for the resource center. |
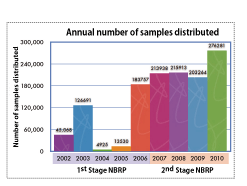
Fig. 1: Number of Samples Distributed Each Year |

Photo: Replication of the collection of mutant strains for all genes. For delivery, 96 deletion mutant strains are simultaneously inoculated on plates by using a pinholder |
|
Is E. coli so different from B. subtilis ?
|
E. coli and B. subtilis are both prokaryotes, but they are significantly different from each other. E. coli normally inhabits the colon, whereas B. subtilis is a soil bacterium that belongs to the same species as B. natto. B. subtilis forms spores under nutrient deprivation. Both species are closely associated with human life, but they exhibit completely different biological life cycles. In fact, the genetic composition of these bacteria indicates that there are only a few genes that are common to both species and that most of the genes are unique to one species or the other (Fig. 2).
Prokaryotes are not so simple that the gene functions of all prokaryotes can be determined by analyzing these 2 bacteria. Nevertheless, prokaryote genome research has progressed because of research conducted on these 2 prokaryotes, as their gene functions have been empirically studied more than those of any other species. Therefore, E. coli and B. subtilis bioresources are indispensible research materials for many prokaryote researchers. |

Fig. 2: Common genes in E. coli and B. subtilis
The number of common genes in B. subtilis is higher than that in E. coli because of the duplication of B. subtilis genes. |
Well-developed collections of protein expression vectors |
| Many researchers who do not necessarily research E. coli itself use E. coli as research material. E. coli is frequently used to express proteins. Multitudes of expression vectors for E. coli have been developed. More than 400 types of vectors, a fraction of the developed vectors, are distributed by our resource center. Readers of this newsletter may be able to find a vector that is useful for their experiments. |
Enhancement of new resources |
| One of our major goals is the enhancement of B. subtilis resources by developing more knockout strains (Table 1). In addition, we have been collecting E. coli and B. subtilis strains with the minimal gene sets and are preparing for the release of these strains. These strains have been developed by eliminating unnecessary gene regions in the genome so that introduced genes can be expressed more efficiently. We have developed both E. coli and B. subtilis strains in which more than 1 Mbp of nucleotides were eliminated from the genome. These strains will soon be available for distribution. We have been developing a resource center that distributes E. coli and B. subtilis bioresources, which are representative species of the gram-negative∗1 and gram-positive∗2 bacteria, respectively. We sincerely hope that the readers will find useful information from our website and use it in their research. |

Table 1: Overview of prokaryote resources |
| ∗1 |
Gram-negative bacteria: Bacteria that stain red or pink-red (negative) on Gram staining (a method to distinguish bacteria into 2 large groups by staining with a dye). Gram-negative bacteria have double-layered thin cell walls and include approximately 80 genera, such as E. coli, Vibrio cholera, Neisseria gonorrhoea, and root nodule bacteria. |
| ∗2 |
Gram-positive bacteria: Bacteria that stain indigo-blue or blue-purple (positive) on Gram staining. Gram-positive bacteria have single-layered thick cell walls without outer membranes and include approximately 60 genera, such as staphylococci, Clostridium botulinum, B. subtilis, and lactobacilli. |
|
|
|
From Internet Explorer 6 to a New Browser |

Ten years have passed since the release of the Internet Explorer 6 (IE6) in 2001. I wonder how many of the readers still use IE6 and I recommend that those readers think about switching over to a new browser.
Microsoft, the developer of IE, opened a campaign site called "The Internet Explorer 6 Countdown" (http://www.ie6countdown.com/) that aims to reduce the number of IE6 users to 1% of the total number of IE users. The website shows the number of IE6 users in the world.
|
|
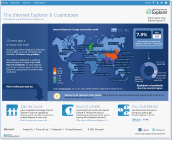
Fig. 1: The Internet Explorer 6 Countdown |
According to the website, as of October 2011, IE6 was still used by an average of 7.9% users across the globe. In Japan, 6.8% of users have IE6, a lower percentage than the global average. However, the percentage of IE6 users is relatively high in East Asia compared to America and Europe.
Update program support for IE6 Service Pack (SP) 3 is scheduled to continue until April 8, 2014. However, the National Information Security Center instructed government agencies to facilitate the transition from IE6 to IE8 in their report, “Activities regarding the transition from old to new browsers.
Reference:http://www.nisc.go.jp/press/pdf/browser_transition_press.pdf (Japanese only)
|

In addition, the new browser is more user-friendly, has higher security than IE6, and runs significantly differently than IE6. Moreover, because supporting IE6 is costly, support for IE6 will soon be terminated, and various services are unavailable in Yahoo! JAPAN and Google to users of IE6.
The new browser has a tab function that prevents computer screens from becoming inundated by many newly opened windows. The new browser can also block users' access to phishing websites or to sites that let users download malicious software. |
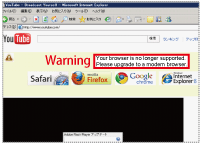 |
Fig. 2:
Attempting to access YouTube (http://www.youtube.com/) with IE6 will prompt the warning message, "Your browser is no longer supported. Please upgrade to a modern browser."
|
|
| |
Windows XP users can upgrade their browser to IE8 using the following procedure:
① First, ensure the operation system of the computer is Windows XP SP2 or SP3.
② Download the file for IE8 from the Windows IE8 for Windows XP website
(http://windows.microsoft.com/en-US/internet-explorer/downloads/ie-8) .
③ Run the downloaded file to install IE8.
|
|
The websites released by our information center will load faster if the new browser is used. Therefore, I highly recommend that IE6 users take this opportunity to switch to the new browser.
|
(Gaku Kimura)
|
|