|
■ Bioresources information is available at the following URLs
|
 |
|
Research and Bioresources <No. 28> Request to deposit genetic materials
Takehide Murata1 • Yuichi Obata2
Gene Engineering Division, RIKEN BioResource Center
(1: Research Scientist 2: Director)
In the National BioResource Project (NBRP), the Japan Agency for Medical Research and Development, Ministry of Education, Culture, Sports, Science and Technology, Gene Engineering Division, RIKEN BioResource Center was selected as a central organization and is responsible for developing Japan’s research foundation. The mission of bioresource projects is to mediate between the developers and users of bioresources, and the mission cannot be accomplished without the understanding and cooperation of the depositors and users of bioresources, i.e., research communities. To promptly provide the latest genetic material developed by foreign and domestic researchers, the Gene Engineering Division has been writing to the authors of papers regarding depositing the genetic resources used in their papers. If you receive such a letter, please deposit your genetic resources at our center. |
Advantages of depositing genetic materials |
Genetic materials can now be easily prepared using polymerase chain reaction or similar techniques. However, to correctly and promptly forward research, using genetic materials published in papers is the most efficient approach. It is the researcher’s responsibility to share the genetic materials with his or her research community and to conduct experiments. Research communities will admire researchers when they quickly provide genetic materials to their fellow researchers. However, researchers’ preparations and provisions of genetic materials become heavy temporal and economic burdens on them and their research may be hindered. When researchers deposit a genetic material at our center, we execute operations for them, such as the amplification and delivery of and paperwork on the genetic material necessary to its provision. Immediately after a paper is published, the number of requests to provide the genetic material used in the paper is highest for the papers of the highest quality. When you decide to submit your paper to a journal, please deposit the genetic material used in your paper at our center. We will take all possible measures so that our center provides genetic material used in your paper immediately after your paper is published.
By depositing your genetic materials at our center, you can prevent those that are kept at your laboratory from being lost or scattered due to unexpected accidents. When researchers deposit their precious genetic materials at our center, the deposited materials can be continuously used by researchers, and these researchers can contribute greatly to the development of Japan’s intellectual foundation. |
Upon the deposition of a genetic material, our center expresses its sincerest appreciation to the depositor. For a genetic material to be cited in a paper or to be used for a commercial purpose, the depositor can add separate conditions, such as a license agreement. Some people believe that a depositor will be annoyed when the deposited genetic material is provided without his or her knowledge of the provision. Therefore, depositors can set a condition that the deposited genetic material can be provided only with the consent of the depositor. However, for genetic materials to be used by many researchers, we ask that depositors set conditions as generously as possible for the deposition and provision of genetic materials. As a matter of course, after depositing a genetic material, the depositor can freely use the material for his or her personal research without restrictions.
For those who have deposited genetic materials at our center, we provide the same type of genetic resources on a free-of-charge basis with our gratitude, the number of which is equivalent to that of the deposited genetic materials (a credit scheme). For example, when you deposit three plasmid clones, you can obtain three plasmid clones of the same type. Only the RIKEN BioResource Center provides genetic materials on a free-of-charge basis equivalent to that of the deposited genetic materials.
For the deposition of genetic materials, please refer to the homepage of Gene Engineering Division at http://dna.brc.riken.jp/en/deposition.html. |
|
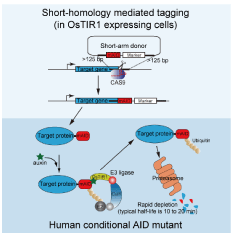
Fig. 1. Decomposition and removal of a target protein using the auxin-inducible degron (AID) system (provided by Associate Professor Masato Kanemaki at the National Institute of Genetics)
By adding auxin to the medium, the decomposition of a target protein can be induced. Namely, mAID is tagged to the gene coding for the target protein by the knock-in method using CRISPR/Cas9. Twenty-one resources used in a paper written by Natsume, T., et al., Cell Reports 15: 210-218 (2016) were provided immediately after the paper was published. The details of the 21 resources are available at
http://dna.brc.riken.jp/ja/gene_analysis/rdb08468ja |
Quality examination of deposited genetic materials |
When deposited genetic materials are examined, the results of the examinations have unfortunately differed from those described in the papers or datasheets in about 10% of the deposited genetic materials. This means that errors are present in approximately 10% of the genetic materials collected by our center or exchanged among researchers.
Our center makes every effort to correct as many of the errors found in genetic materials as possible and to provide genuine resources by eliminating the genetic materials in which errors cannot be corrected. Through this process, the reproducibility and efficiency of the research results can be secured.
|
We have determined quality control standards according to the type of genetic material
(http://dna.brc.riken.jp/ja/resource) .
For example, the following matters have been examined for plasmid and cosmid clones: (1) growth and antibiotic resistance of the recombinant bacteria in the culture conditions specified by the depositor/developer, (2) restriction enzyme mapping, and (3) nucleotide sequencing around a vector-insert junction area and portions of vector sequences.
The examination results are shown in the web catalog. The electrophoresis profile can be used as a control in an examination to confirm whether the user has received the correct genetic material after amplifying it. Base sequences are opened to the public not only as character data, but also as chromatogram with handwritten annotations, and they are useful as references when the provided genetic materials are further customized and used.
Information on the structure of a resource obtained in an examination (such as the existence of a restriction enzyme recognition sequence, data on an experiment which has previously been performed by the depositor/developer, papers in which genetic materials were established, and papers for which genetic materials were provided for the authors) is appended to the catalog as supplemental information to prove the validity of the corresponding genetic material.
Each genetic material has been registered in PubMed LinkOut※) through the Research Resource Circulation, a database in the NBRP. Researchers can easily access genetic materials that have been used in papers of interest.
We would greatly appreciate your information on depositing your genetic materials at our center. |
|
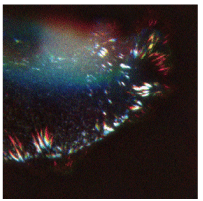
Fig. 2. Movement of focal adhesion (FA) in a MDCKII cell observed using a bright luminescent protein Nano lantern (NL) obtained by accumulating sequence photographs for four hours (provided by Dr. Yasushi Okada of the RKEN Quantitative Biology Center).
FA moved from the location indicated in blue to the location indicated in red. White (when these two colors overlapped each other) indicates stationary FA. A total of 51 clones, used in Takai, A. et al., Proc. Natl. Acad. Sci. U. S. A., 112: 4352‒4356 (2015), were deposited at our center. These included clones by which intercellular fine structures, such as nuclei, lysosomes, and cytoskeletal fibers can be observed by yellow-green, blue-green, and orange NLs, respectively, in addition to the Vinculin-Orange Nano-lantern shown in this figure, and clones in which the promotor of each of Nanog, Oct4, and Sox2 genes is connected with NL. The details of these resources are available at http://dna.brc.riken.jp/ja/nano_lantern |
| ※ |
For PubMed LinkOut, please refer to "Ongoing Column No. 89" in BioResource Now! Vol. 10 No. 6 (June 2014). |
|
|
|
|
Linux tools can now be run on Windows 10 ! Part 1 |

Various software tools are used in research involving the use of next-generation sequencers (NGS). Many of these are command-line tools and often cannot be installed on Windows. In the past, various virtualization techniques including Cygwin, VMware, and VirtualBox were used to run these tools on Windows. However, it has now become possible to run Linux tools on Windows using "Bash on Ubuntu on Windows" that was released as part of the Windows 10 Anniversary Update on August 2, 2016. This article explains how to install this function.
※The functionality introduced in this article requires that you are running Windows 10 with Anniversary Update already installed. Other operating systems, including Windows 8.1 and others, cannot be used.
※ • Bash on Ubuntu on Windows is still in its beta stage. Please use it at your own risk.

Installing Bash on Ubuntu on Windows
"Developer Mode" on Windows 10. Click on the Start button and select [Settings]-[For developers] and select "Developer mode" (Fig. 1).

Fig. 1. Developer mode
You must restart Windows after enabling developer mode. You should shut down all applications before restarting.。
Next, right-click on the Start button and select [Control Panel]−[Programs]−[Turn Windows features on or off], select "Windows Subsystems for Linux (Beta)" (Fig. 2), and then restart Windows again
After restarting, open the command prompt, type "bash," then hit the [Enter] key. Press "y" when prompted to accept the license agreement to begin the installation (Fig. 3).
|
|
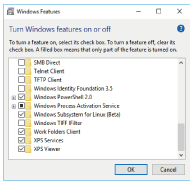
Fig. 2. Enabling Windows Subsystem for Linux (Beta)
|
|
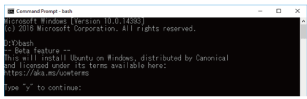
Fig. 3. Installing bash
|
During the course of the installation, you will be asked to enter a username and a password for use with Bash on Ubuntu on Windows (this username is different from that of your Windows logon account).
After the installation is complete, either enter "bash" from the command prompt, or choose [Bash on Ubuntu on Windows] from the Start menu to launch the tool.
In the next issue, I shall introduce how to install the "TOPHAT" software.
(Shunsuke Maeda) |
|
|