|
 |
■ Bioresources information is available at the following URLs
|
 |
|
Introduction to Resource Center <No.38> National BioResource Project C. elegans
Shohei Mitani, Professor, School of Medicine, Tokyo Women's Medical University
Time goes by so fast; 10 years have already passed since we began operations as the core institution of the National BioResource Project (NBRP) C. elegans. The history of research on Caenorhabditis elegans probably stretches back to the 1960s when Dr. S. Brenner started using C. elegans in his research in Cambridge, UK, and therefore, the 10-year duration is not negligible. During that time, we have continuously been collecting, preserving, and distributing C. elegans resources as the core institution of the NBRP C. elegans at the Tokyo Women’s Medical University, Japan. As a result, the number of publications in which our mutant resources were used has consistently increased. In fact, it may even be regarded as magical that we have successfully advanced our project since its inception. I feel this way because we have not changed the overall framework of collecting, preserving, and distributing the deletion mutants, although we have occasionally applied new technologies. It is amazing to see that the productivity could be maintained simply by repeating the same tasks for many years in a scientific world; however, this is to be attributed to the kind understanding and support of many people.
It would be important to develop a system that improves the relationship between the NBRP in Japan and research communities in the world to promote our bioresource activities. For this purpose, we first need to familiarize C. elegans researchers with our NBRP activities. Almost all the C. elegans researchers use WormBase (http://www.wormbase.org/), an integrated database in which large quantities of data for each gene as well as other data obtained in the past have been archived and in which any known information regarding C. elegans can be found. We were requested by the WormBase team to provide a copy of the NBRP data, and we uploaded the data to the NBRP C. elegans site hosted at the Sanger Institute, where the curators of the mutant section of the WormBase work. WormBase collects so much information that it takes 2–3 months to standardize the format of all the data available at a certain point and integrate them into the new version of the database, which is availed of by every C. elegans researcher and is therefore considered valuable. For example, upon searching for a gene of interest, one can find the gene structure displayed on the first screen, as well as the deleted regions of any deletion mutants (Fig. 1). Anyone interested in the gene would consider the phenotypic information of the loss of the gene function to be an important clue for their research, and would therefore be likely to order the mutant strain.
Meanwhile, although data obtained by other researchers using bioresources are important, no experiment can be conducted without obtaining bioresources. This will be the motivation for researchers to be guided to the NBRP website, http://shigen.lab.nig.ac.jp/c.elegans/. If the mutant strain is listed in the site, one can obtain original information regarding the mutant by clicking on the link to the mutant allele in WormBase. By providing mutual links in both the NBRP and WormBase websites, we have developed a framework to guide researchers who browse the WormBase and find mutants of their interest to the NBRP website (Fig. 2). |
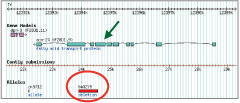
図 1:Fig. 1: Deleted region (encircled) and gene structure (↙) of a deletion mutant |
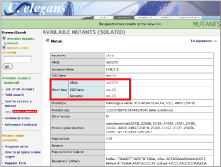
Fig. 2: Link from the NBRP site to the WormBase site |
At the same time, a server at the National Institute of Genetics provides a website specifically prepared for the NBRP database, where core institutions can always upload new data. Core institutions upload new mutant information to both the NBRP and the WormBase websites simultaneously, and the NBRP website immediately releases the information. On the other hand, the information in the WormBase site can be accessed by researchers only after 2–3 months. Therefore, many users who frequently access the NBRP website and are aware of the time difference will bookmark the site and visit the NBRP site frequently and preferentially. By doing this, users will be more likely to find mutant strains of their interest. In addition, we collect and preserve approximately 500 new deletion mutants per year. If users who are well acquainted with the above-mentioned websites cannot find mutant strains of their interest in WormBase, they can submit a request for screening the mutants. By doing this, users will be accustomed to accessing the NBRP website, searching by gene name, and waiting for mutants of their interest to be isolated and released. This is supposedly how the website is used by advanced users, since newly released mutant strains are sometimes requested for distribution immediately after their release.
It is extremely important for researchers to obtain new mutants and elucidate gene functions of interest as soon as possible. At the same time, we also assume that there is a need to analyze novel compounds using C. elegans, which has well-documented fundamental information and has been used in research for half a century as a model organism. Based on such information, the functions of several C. elegans genes have been elucidated using RNAi, mutants, and transgenic strains. We wonder what would be the next to be elucidated. |
We isolated a deletion mutant of C. elegans, analyzed its phenotype, and identified a unique gene mutant called acs-20 (E. Kage-Nakadai, et al., PLoS One, 2010) (Fig. 1–3). This mutant strain is permeable to small-molecule compounds. The biosynthesis of lipids that form the epidermal barrier appears to be affected in the mutant. Since no other critical phenotype has been identified under a standard culture condition, the mutant strain can be used to analyze whether phenotypes of interest will be changed in the background of acs-20 mutation. This mutant has been requested for distribution by many laboratories after the publication of the strain.
C. elegans can be cultured in liquid medium in 96-well plates and can therefore be used for screening. I expect that the website will be a tool for researchers to investigate the mechanisms of action of many compounds in C. elegans, in addition to the accumulated gene information. I also believe that the NBRP C. elegans will contribute not only to the accumulation of data by analyzing new gene functions with new mutants but also to provide an analysis tool for elucidating the mechanisms of novel compounds. |
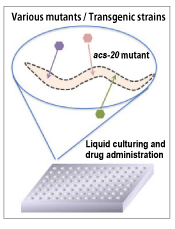
Fig. 3: Gene mutant (acs-20) |
|
|
Functions in Microsoft Office SmartArt |

When we prepare presentation slides with figures, many would feel it troublesome to paste shapes and connect them with lines. In this issue, I will introduce SmartArt, which can be used to quickly prepare flowcharts and hierarchical structures in PowerPoint, Excel, or Word of Microsoft Office 2007 or later versions for Windows, and in Microsoft Office 2008 or later versions for Mac.
|
| |
Prepare Shapes using SmartArt
First, run PowerPoint, Excel, or Word application and select SmartArt from the insert tab of the menu bar (Fig. 1-A). Once the selection dialog of SmartArt graphics is displayed as shown in Fig. 1, select a template that suits your needs.
|
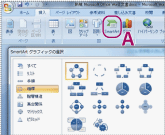
Fig. 1: Selection of a shape |
|
| |
Texts can directly be edited within the selected template shape (Fig. 2-A), or by pressing the text window (Fig. 2-B), to display the text input box (Fig. 2-C).
|
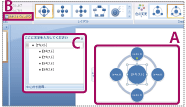
Fig. 2: Editing text |
|
| |
In addition, elements displayed in the figure can be added or deleted by selecting the target element in the text input box (Fig. 3-A) and hitting the Delete key to delete, or the Enter key to add an element. The positions of the remaining elements will then be automatically adjusted after adding or deleting elements (Fig. 3-B).
|
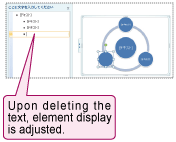
Fig. 3: Deleting an element. |
|
| |
How can we change the layout ?
By selecting other layouts from the design tab in the menu bar, the layout of the figure alone can be changed without disturbing the text (Fig. 4).
|
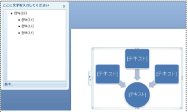
Fig. 4: Changing the layout |
|
| |
Although I have introduced SmartArt in this issue, the PowerPoint, Excel, and Word applications of Microsoft Office 2010 feature more than 50 new templates, to which images can be attached. This function has rarely been seen before.
In addition, a subset of SmartArt templates for Microsoft Office 2010 can be downloaded from the Microsoft Office website and added, but this is restricted to users of Microsoft Office 2007 for Windows alone. In order to add templates, download the templates of your interest from the URL of the Microsoft Office website (in English) below and select download button in the detailed screen.
Download URL:
http://office.microsoft.com/en-us/templates/CL101829681.aspx?CTT=5&origin=HA010211779
∗ Those who use browsers other than Internet Explorer should unzip the downloaded file and move the file, [~.glox], to the address,
[C:\Users\\AppData\Roaming\Microsoft\Templates\SmartArt Graphics] (in Windows 7).
This will complete adding the templates, which will be displayed in the SmartArt of PowerPoint, Excel, and Word applications. Although the templates can be downloaded from the Microsoft Office website in Japanese, the number of downloadable templates is lesser than those on the English website.
|
|
--------------------------------------
It is recommended that those who need to prepare slides avail of the various figures prepared, which are categorized into lists, procedures, circulation diagrams, hierarchical structures, group relationships, matrices, and pyramids.
|
(HIroki Watanabe)
|
|